Quick custom comorbidity maps are helpful
Jack O. Wasey
2020-06-08
Source:vignettes/custom-maps.Rmd
custom-maps.RmdMany problems with clinical data are best solved using the standard and widely validated comorbidities, mapped to subsets of ICD-9 or ICD-10 codes, as published by authors including Elixhauser, Quan, Deyo and the AHRQ. It is also common to have a pre-defined specific set of ICD codes. Examples of this include:
- Studying sub-types of a common comorbidity, such as obesity: although obesity is in the AHRQ and Elixhauser mappings, there is no granularity.
- Studying a particular disease: most diseases are not in the standard comorbidity maps. To distinguish patients having or not having a particular set of diseases is usually not possible using the standard comorbidity maps. E.g. Which patients have eczema? Which emergency room patients presented with a sexual health problem?
icd has a simple mechanism to use custom category-ICD maps. A comorbidity map is a list of character vectors. Each list must be named to reflect the ICD codes it contains; the character vector contains the ICD codes themselves.
Let’s take a look at the first few items in the Charlson map from Quan and Deyo:
print(icd::icd10_map_quan_deyo, n_comorbidities = 3, n_codes = 8) #> Showing first 3 comorbidities, and first 8 of each. #> $MI #> [1] "I21" "I210" "I2101" "I2102" "I2109" "I211" "I2111" "I2119" #> #> $CHF #> [1] "I099" "I110" "I130" "I132" "I255" "I420" "I425" "I426" #> #> $PVD #> [1] "I70" "I700" "I701" "I702" "I7020" "I70201" "I70202" "I70203" #> #> ...
Obesity example
Now we can make our own comorbidity map, and apply it to a question on ‘Stackoverflow’.
library(icd) obesity_map <- list( "1" = "E663", "2" = c("E669", "E668", "E6609"), "3" = "E661", "4" = "E662") obesity <- data.frame( ICD.10.Code = c("E6601", "E663", "E663", "E6609"), Encounter.ID = c("408773", "542207", "358741", "342534") ) custom_map_result <- icd::icd10_comorbid( obesity, map = obesity_map) custom_map_result #> 1 2 3 4 #> 408773 FALSE FALSE FALSE FALSE #> 542207 TRUE FALSE FALSE FALSE #> 358741 TRUE FALSE FALSE FALSE #> 342534 FALSE TRUE FALSE FALSE # finally, format as requested by the user apply(custom_map_result, 1, function(x) { if (!any(x)) 4 else which(x)[1] }) #> 408773 542207 358741 342534 #> 4 1 1 2 # see also: icd::icd10_map_ahrq$Obesity #> [1] "E6601" "E6609" "E661" "E662" "E668" "E669" "O99210" "O99211" #> [9] "O99212" "O99213" "O99214" "O99215" "R939" "Z6830" "Z6831" "Z6832" #> [17] "Z6833" "Z6834" "Z6835" "Z6836" "Z6837" "Z6838" "Z6839" "Z6841" #> [25] "Z6842" "Z6843" "Z6844" "Z6845" "Z6854" icd::icd10_map_quan_elix$Obesity #> [1] "E66" "E660" "E6601" "E6609" "E661" "E662" "E663" "E668" "E669" icd::icd10_comorbid_quan_elix( obesity, return_df = TRUE)["Obesity"] #> Obesity #> 1 TRUE #> 2 TRUE #> 3 TRUE #> 4 TRUE
Maps with ranges of codes
Sometimes there are a large number of ICD codes, and they can be defined more succinctly with ranges, then by specifying every single code. In addition, as new codes are added to ICD-10-CM, (especially ICD-10-CM – it moves much faster than ICD-10 from the WHO) having specific hard-coded ICD-10 codes will miss closely related codes in the future. Using the ranges functions from icd helps with both these problems.
Again, using a Stackoverflow question:
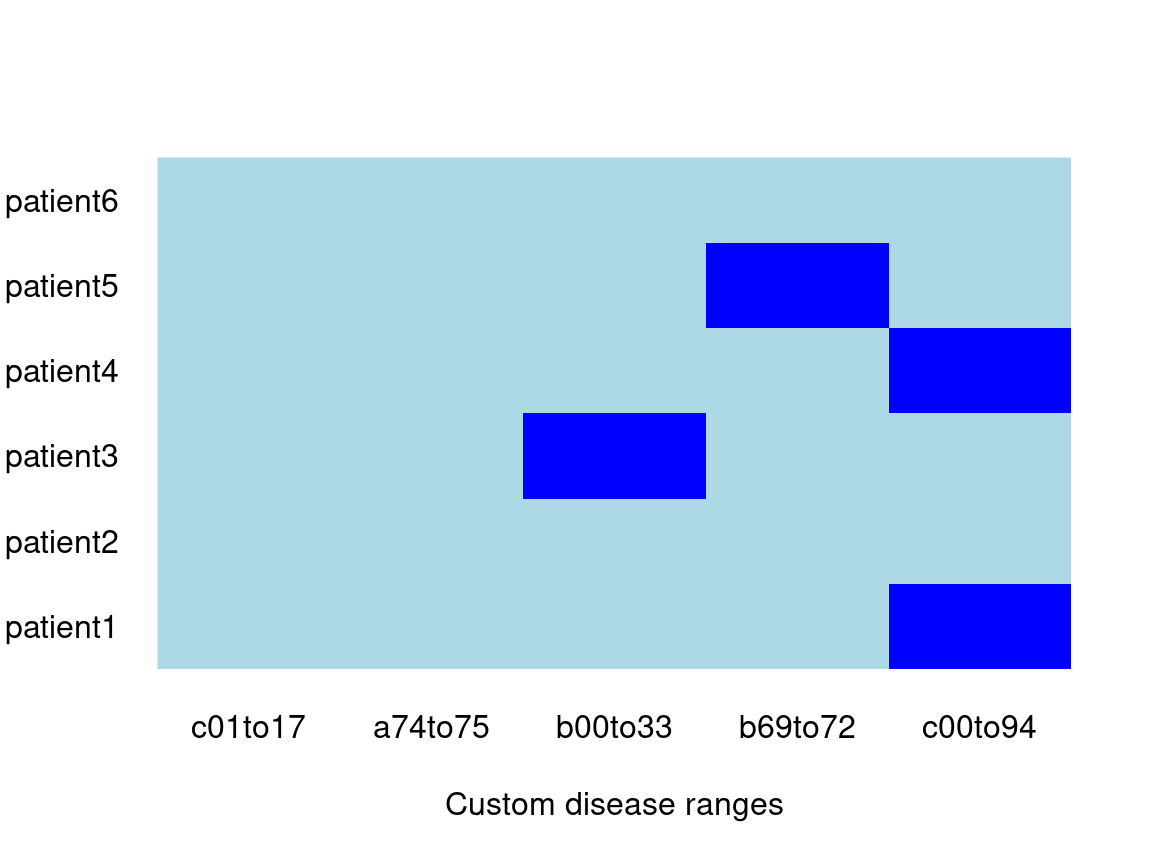
library(icd) diagnoses <- c("C349", "A219", "B003", "C509", "B700", "A090") one_pt <- data.frame(id = rep("patient1", length(diagnoses)), diagnoses) dif_pt <- data.frame(id = paste0("patient", seq_along(diagnoses)), diagnoses) my_map <- list(c01to17 = expand_range("C01", "C17"), a74to75 = expand_range("A748", "A759"), b00to33 = expand_range("B001", "B331"), b69to72 = expand_range("B69", "B72"), c00to94 = expand_range("C000", "C942")) # optionally use as.comorbidity_map which ensures it is valid, and let's it # print more pleasantly my_map <- as.comorbidity_map(my_map) print(my_map) #> Showing first 7 comorbidities, and first 7 of each. #> $c01to17 #> [1] "C01" "C02" "C020" "C021" "C022" "C023" "C024" #> #> $a74to75 #> [1] "A748" "A7481" "A7489" "A749" "A75" "A750" "A751" #> #> $b00to33 #> [1] "B001" "B002" "B003" "B004" "B005" "B0050" "B0051" #> #> $b69to72 #> [1] "B69" "B690" "B691" "B698" "B6981" "B6989" "B699" #> #> $c00to94 #> [1] "C000" "C001" "C002" "C003" "C004" "C005" "C006" icd::comorbid(one_pt, map = my_map) #> c01to17 a74to75 b00to33 b69to72 c00to94 #> patient1 FALSE FALSE TRUE TRUE TRUE (six_pts_cmb <- icd::comorbid(dif_pt, map = my_map)) #> c01to17 a74to75 b00to33 b69to72 c00to94 #> patient1 FALSE FALSE FALSE FALSE TRUE #> patient2 FALSE FALSE FALSE FALSE FALSE #> patient3 FALSE FALSE TRUE FALSE FALSE #> patient4 FALSE FALSE FALSE FALSE TRUE #> patient5 FALSE FALSE FALSE TRUE FALSE #> patient6 FALSE FALSE FALSE FALSE FALSE